Instance-based learning
In this project, you will implement an efficient instance-based learner.
- Read (or skim) Section 2.2 of the book.
- Implement a k-nn instance-based learner. To help you get started, here is some inefficient code to
find the k-nearest neighbors of a point. (Disclaimer, this code has not been tested. You may need to
debug it.)
bool my_comparator(const pair<size_t,double>& a, const pair<size_t,double>& b) { return a.second < b.second; } void find_neighbors_by_brute_force(const Vec& point, const Matrix& data, size_t k, vector<size_t>& outIndexes) { vector< pair<size_t,double> > index_dist; for(size_t i = 0; i < data.rows(); i++) { double dist = compute_distance(point, data[i]); index_dist.push_back( pair<size_t,double>(i, dist) ); } std::sort(index_dist.begin(), index_dist.end(), my_comparator); outIndexes.resize(k); for(size_t i = 0; i < k; i++) outIndexes[i] = index_dist[i].first; }For continuous values, use Mahalanobis distance. For categorical values, use Hamming distance. - Please make sure your instance-based learner passes the following unit tests:
First, train on this data,@relation knn_train_features @attribute x real @attribute y real @data 0.9,3 2,2.01 4,2 2,2.5 0,0 3,0 1,5 0,2
@relation knn_train_labels @attribute class {a,b} @data a a a b a a a athen test on this data using k=3, and a weight-exponent of 1 (linear weighting):@relation knn_test_features @attribute x real @attribute y real @data 2,3 2,4 0,0
@relation knn_test_labels @attribute class {a,b} @data b a aIt should correctly classify all three points. If not, try doing it by hand on a piece of graph paper. Then, see where your code does something different.
Here is another test. First, train on this data, (the last two columns are labels),@relation knn_train_features @attribute f1 {a,b} @attribute f2 {a,b} @attribute f3 {a,b} @attribute f4 {a,b} @attribute f5 {a,b} @data a,b,a,a,a a,a,a,b,b b,b,a,b,a b,b,b,b,b b,a,a,b,b a,a,a,b,a
@relation knn_train_labels @attribute l1 real @attribute l2 real @data 2,3 2.5,2.5 37,14 29,19 -71,-11 3,2
then test on this data with k=3,@relation knn_test_features @attribute f1 {a,b} @attribute f2 {a,b} @attribute f3 {a,b} @attribute f4 {a,b} @attribute f5 {a,b} @data a,a,a,a,aThe 3-nearest neighbors should be {0,1,5}. The distances should be {1,2,1}. The predicted label vector should be {(1/1*2+1/2*2.5+1/1*3)/(1/1+1/2+1/1) = 2.5, (1/1*3+1/2*2.5+1/1*2)/(1/1+1/2+1/1) = 2.5}. - Here is some medical data. Train your k-nn instance-based learning using the training features and labels. Classify the test features. Report the number of misclassifications with 3 neighbors using linear weighting. (The correct answer is greater than 0 and less than 100.) A good way to make sure you do it right is to not even load the test labels until after you have made all of your predictions.
- Implement a kd-tree that can be used to efficiently find the k-nearest neighbors of a vector.
Each leaf node in your tree should store the indexes (not the actual vectors) that it encloses.
Here are some example data structures that might be helpful for getting started:
class KdNode { virtual ~KdNode() {} virtual bool isLeaf() = 0; }; class KdNodeInterior : public KdNode { size_t column; double value; KdNode* less_than; KdNode* greater_or_equal; virtual ~KdNodeInterior() { delete(less_than); delete(greater_or_equal); } virtual bool isLeaf() { return false; }; }; class KdNodeLeaf : public KdNode { vector<size_t> m_pointIndexes; virtual ~KdNodeLeaf() {} virtual bool isLeaf() { return true; }; }; class KdTree { Matrix m_points; KdNode* m_pRoot; KdTree(Matrix& points) { m_points.copy(points); vector<size_t> indexes; for(size_t i = 0; i < points.rows(); i++) indexes.push_back(i); m_pRoot = buildKdTree(indexes); } ~KdTree() { delete(m_pRoot); } void buildKdTree(vector<size_t>& indexes); void findNeighbors(size_t k, const Vec& point, vector<size_t>& outNeighborIndexes); };
Here is some pseudocode for building a kd-tree:Let P be a set of point indexes. if P contains fewer than 8 (or so) point indexes, then return a leaf node containing all of P. else Measure the mean and deviation in each dimension. Let i be the dimension with the largest deviation. Divide P into two sets by dividing at the mean in attribute i. Recursively build the two child nodes. return an interior node that joins those two child nodes.
Here is some pseudocode for finding neighbors using a kd-tree:Let p be a point for which we want to find the k-nearest neighbors. Let s be an initially empty priority queue of point indexes, sorted by distance to p. Let q be an initially empty priority queue of KdNodes, sorted by minimum distance to p. Add the root node of the kd-tree to q. while q has at least one node in it, do: pop n from q if n is a leaf node, then: add all the points in n to s else if the distance from p to n is greater than the kth neighbor in s, then break add both child nodes of n to q return the k-nearest points in s
A good way to debug your kd tree is to fill an n-by-2 matrix with random values. Next, find the smallest value for n where your kd tree gives different results from the brute-force method. Finally, plot the random points on a piece of graph paper, then step through your code and see where it does something wrong. After you get it working, crank up the value of n, until you have convinced yourself that your kd tree is bullet-proof. - Measure and report how much speed-up your kd-tree gives relative to the brute force approach.
For Java, nanotime is good for measuring time. For C++, here is some code:
double seconds() { #ifdef WINDOWS return (double)GetTickCount() * 1e-3; #else struct timeval tp; gettimeofday(&tp, NULL); return ((double)tp.tv_sec + (double)tp.tv_usec * 1e-6); #endif } - I don't want to wait to see your code do it the slow brute-force way, so please turn in your code in a state that only uses the kd-tree. (Just print the measured speed-up as hard-coded output.) Also, please keep your output simple. (I don't need to see all your debugging spew.) Then, submit a zip file containing your code and the data it needs to work. (Do not submit any generated binary files.) Make sure your zip file contains a file named build.bash (or build.bat) that builds your code.
Q and A
- How does one measure the distance between a point and a KdNode?
Consider this figure:
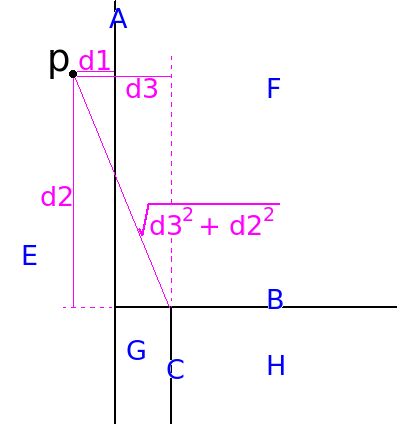
The Kd tree represented in this figure contains 3 interior nodes (A, B, and C) and four leaf nodes (E,F,G, and H). Suppose we want to find the distance between p and H. In order to find H, we must traverse the tree from A->B->C->H.
Let v be a vector of size 2 (because there are two dimensions in this example). It stores the squared distance in each dimension. Initialize v with zeros, such that v=[0, 0].
The distance from p to A is sqrt(0 + 0). This is correct because A encloses all points.
When we visit B, we update v to be [d1*d1, 0]. The distance from p to B is sqrt(d1*d1 + 0*0).
When we visit C, we update v to be [d1*d1, d2*d2]. The distance from p to C is sqrt(d1*d1 + d2*d2).
When we visit H, we update v to be [d3*d3, d2*d2]. The distance from p to H is sqrt(d3*d3 + d2*d2). (Since A and C both divide in the same dimension, We stomp over the old value in that dimension.)
The distance from p to E, should be 0. The distance from p to F should be d1. - How does one measure the deviation of a categorical attribute?
Deviation is a measurement for continuous attributes. A similar metric for categorical attributes is normalized entropy:function computeEntropy(v) Let k be the number of categories. Let n be the number of samples. double h = 0.0; for i from 0 to k-1 int c = 0; for j from 0 to n-1 if v[j] == i c++; h -= c / n * log(c / n) / log(k); return h;
Normalized entropy is not really the same thing as deviation, but it has several similar properties. It always falls between 0 and 1, and larger values indicate that splitting on this attribute will provide more benefit for the Kd-tree. Wouldn't it be like comparing apples to oranges if we compare deviations of normalized attributes against normalized entropies? Yes. However, if you choose to divide on a sub-optimal attribute, that will only make the kd-tree slower. It should not make the kd-tree yield incorrect results. - When the Kd-tree divides on a categorical attribute, do we need more than two child nodes?
No. For categorical attributes, I would compare for equality, not inequality. Example: If value == "potato", then go to left child, else go to right child.